Image Analysis
Spending quality time inspecting your data is critical for hypothesis and analysis pipeline formation. Each microscope manufacturer has different proprietary file formats (ex. .lif for Leica, .czi for Zeiss) that store multidimensional (x, y, z, t, channels, etc.) image data along with image metadata (microscope hardware settings like laser power, filters, voxel size) in uncompressed or lossless compressed form. It’s a good idea, if you have storage capability, to retain your image in this type of format to keep the metadata which are important for reproducibility. However, it may not be obvious how to go about opening these files on your own computer for further visualization and analysis. There are a few options.
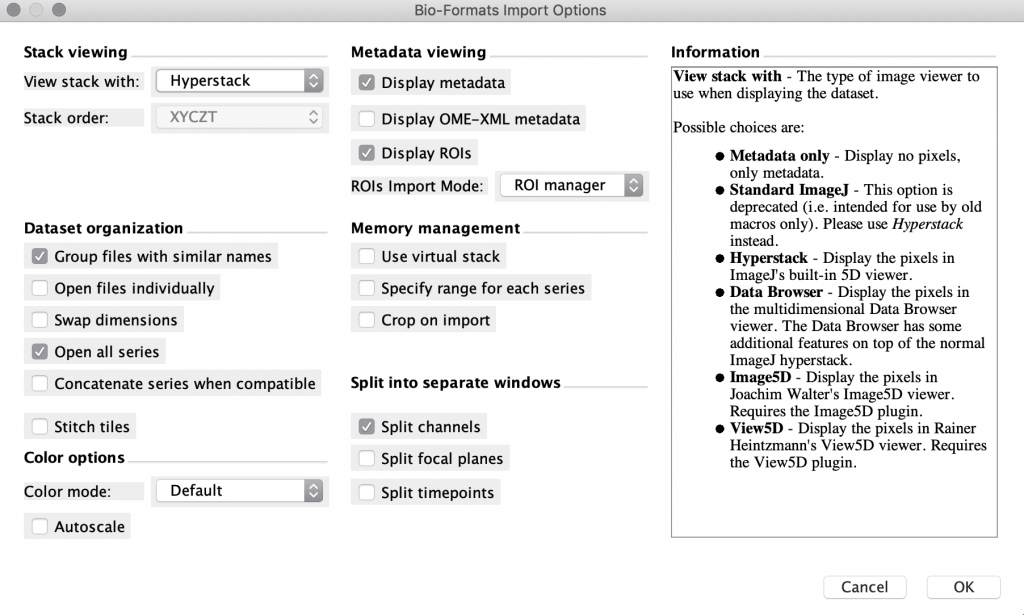
Open the file in FIJI. FIJI (is just ImageJ) is a powerful open-source, free software tool that can open most proprietary file formats and is available for Mac, Linux, and Windows. If you drag your file onto the FIJI status bar, it will use the BioFormats library to open your images as well as display the metadata. Once your data is in FIJI, you can perform 3D visualization and processing of your images and save them in a variety of other formats (eg. .tif, avi, .gif). Sometimes this approach works smoothly, and other times there may be some issues getting the import to work properly. Image.sc is an active discussion forum where you can see if anyone has had similar issues and find potential solutions.
Convert to Imaris format and use the MIC Imaris license. The MIC has an image analysis workstation that has an installation of Imaris, a large-data optimized commercial image analysis software. Imaris converts proprietary microscope image files to .ims files which can be further processed within the software. Commonly used analysis tools include measurement, annotation, object detection, and filament tracing. You can use this software on the workstation at the MIC for $1/hr (max of $8/day), or you can check out a satellite license to use on your own machine for $35/week.
Open the file in free proprietary “Viewer software. Some microscope manufacturers have a free “viewer” version of their image handling software that allows you to do very basic viewing and annotations of their proprietary files. One example is Zen Lite for Zeiss .czi files (Windows only), LAS X Core for Leica files (Windows only), and Imaris Viewer for .ims files (Windows and Mac).
Export as .tif file. When you acquire the images, you can export them as .tif files, but be sure to record the experimental metadata in a secure place. These images can be opened by most image viewing and editing software (eg. FIJI, gimp, photoshop).
Out of memory? In some cases, you may have image data that is too large to fit in the memory (RAM) of your computer, in which case you need either 1) to open the image in FIJI as a “virtual stack” which means only the actively viewed image is loaded into memory from the hard disk rather than the entire stack being loaded into memory, 2) to use a visualization tool that is able to make some smart adjustments to the data storage architecture, caching and loading strategies (eg. FIJI’s BigDataViewer), or 3) to utilize a workstation with larger memory like the Imaris workstation at the MIC which has 64 GB memory, or 4) to use the TAMU HPRC’s interactive portal sessions to access FIJI on a node with a large amount of memory. Please be aware that performing any processing on the data you’re visualizing will likely further increase the memory requirements. For assistance utilizing any of these approaches, please contact Dr. Holly Gibbs (hgibbs@tamu.edu).
It can be difficult to observe specimens with very weak fluorescence or those that photobleach easily using a confocal microscope. Standard epifluorescence imaging is often a method of choice for these objects. Deconvolution is a computational image restoration technique that can remove the out-of-focus blur typical for epifluorescence images and improve both lateral and axial resolution. Deconvolution is best suited for relatively thin specimens (microorganisms, single cells, tissue sections) where it can bring significant improvement in resolution and contrast.
The MIC offers AutoQuant X software with several processing algorithm including the iterative blind deconvolution for wide-field fluorescence images. This software can be used in our computer lab free of charge.
For more information on deconvolution and for help with image acquisition for deconvolution
please contact Dr. Stanislav Vitha, vitha@tamu.edu.
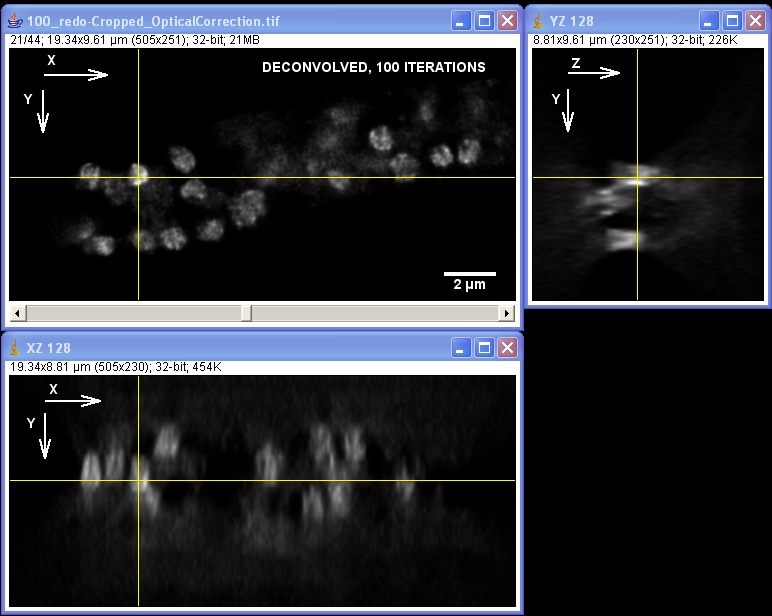
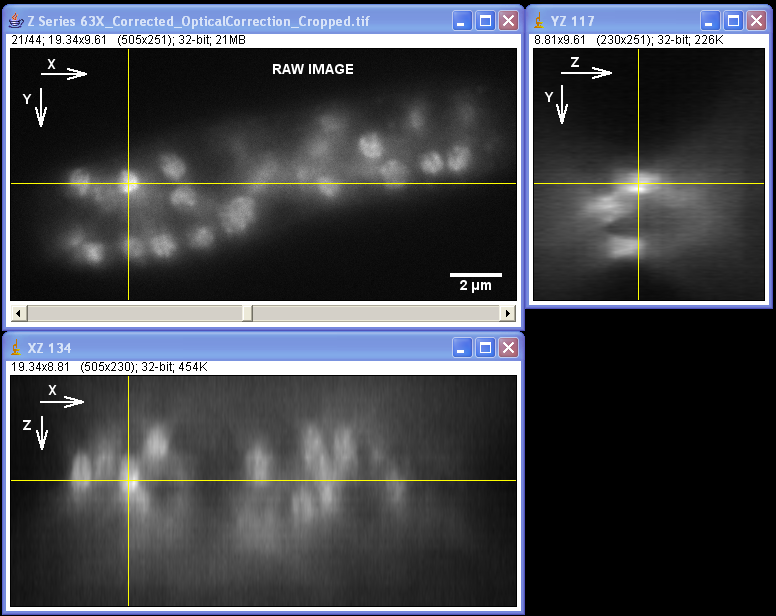
Specimen courtesy of Dr. Bell-Pedersen (Department of Biology).
The Zeiss Axiophot microscope equipped with a Plan Neofluoar 100x/1.3 oil-immersion objective and a Coolsnap cf camera was used to create a z-stack of GFP fluorescence images with z-step of 0.2 um. The raw image stack was processed with AutoDeblur X software (Media Cybernetics) using 100 iterations of blind deconvolution algorithm, taking into account spherical aberration due to refractive index mismatch. The raw and deconvolved image stacks were then opened in ImageJ software to generate orthogonal views of the image stacks. Note: the large amount of out-of-focus blur in the raw image as well as the improvement in contrast and resolvable detail in the deconvolved dataset. Image data was acquired by Laura Short (Department of Anthropology) during the Spring 2009 Light Microscopy course offered by the MIC. Image processing by Stanislav Vitha, MIC.
Software Available with the Instruments
- Optical Microscopes
- Z.1 Lightsheet Microscope–
- Zen Black Acquisition Software acquisition with limited image visualization and processing capabilities.
- Leica SP8–
- LAS X acquisition software modules for multi-dimensional, STED, and FLIM imaging, LIGHTNING deconvolution, 3D image analysis and processing.
- PC with LAS X Core, LAS X FLIM free viewers and ImageJ/FIJI freeware for offline review of images is available in our computer room.
- Olympus Confocal–
- FV10-AFS software with multi-dimensional acquisition.
- PC with full version of FV10-AFS software for off-line analysis of confocal datasets is available in our computer room.
- Leica DM6B–
- LAS X software for multi-channel, time lapse, Z-stack image acquisition, large area scanning, image stitching, and autofocus.
- FIJI freeware for image processing: https://webshare.leica-microsystems.com/latest/core/widefield
- Zeiss Axiophot–
- Micro-Manager software is free software for image acquisition control.
- Z.1 Lightsheet Microscope–
- Transmission Electron Microscopes
- FEI G2 F20 ST TEM Materials–
- Support PC set-up with Inspect 3D for tomographic reconstructions
- Xplore3D for automated tomographic tilt series acquisition in TEM or STEM mode
- FEI F20 Cryo TEM–
- Support PC set-up with Inspect 3D for tomographic reconstructions
- JEOL 1200–
- FEI G2 F20 ST TEM Materials–
- Scanning Electron Microscopes
- Quanta–
- Vega–
- Tescan acquisition software for image acquisition, image processing, measurement, and image stitching
- Bruker Alicona MeX software for 3D surface profilometry from stereo images. **Check-out required** Contact Stanislav Vitha
- Cryo FIB SEM –Auto Slice and View 4.0 (AS & V4) software acquires high-resolution data by milling serial sections (slices) of a specimen with a focused ion beam and then imaging and/or mapping each slice generating a large stack of images.
Software Packages Available at the MIC
- Imaris for image analysis useful for manual and semi-automated spot detection, filament tracing, and regional segmentation in 3D fluorescence images. The MIC license can be utilized on site with a provided analysis workstation. Contact Holly Gibbs to arrange training on Imaris.
- NanoMegas ASTAR is available on the F20 Materials TEM computer workstation designed for analysis of crystallographic phase and orientation for individual grains.
- Amira for analysis and 3D processing, analysis of EM and optical microscopy images, 3D deconvolution, reconstruction, and segmentation. The stack of images and their parameters are seamlessly read from the data headers of the images with Amira and Avizo software package thus allowing the user to generate 3D electron density maps of the specimen.
- Aivia– Automatic Artificial Intelligence software package Aivia offers the users for quick and easy segmentation results.
- Bruker Alicona MeX software https://www.alicona.com/products/mex/ for 3D surface texture analysis, roughness analysis from SEM stereo images **Check-out required** Contact Stanislav Vitha
- LAS X software with FLIM module: Contact Stanislav Vitha
Software Available to Use on Your Personal Computer
- Image J http://rsb.info.nih.gov/ij
- FIJI https://fiji.sc
- FLIMJ plugin for ImageJ/FIJI https://imagej.net/FLIMJ for analysis of Fluorescence Lifetime data from the Leica SP8 system.
- SimFCS https://www.lfd.uci.edu/globals/ Software for fluorescence image analysis, visualization, simulation, and acquisition. Fluoresence Correlation Spectroscopy (FCS), Fluorescence Lifetime Imaging (FLIM) and Phasor plots, Förster Resonance Energy Transfer (FRET), Generalized Polarization (GP) and Spectral Phasors, Number and Brightness (N&B), Photon Counting Histogram (PCH), Raster and Spatio-temporal Image Correlation Spectroscopy (RICS and STICS), Single Particle and Modulation Tracking (SPT, MT), Image Mean Square Displacement (iMSD),Pair correlation function (pCF).
- Olympus Fluoview Confocal Image Viewer https://www.olympus-lifescience.com/en/support/downloads/#!dlOpen=%23detail847252002 **our Serial Number is 5L10**
- Las X Core wide-field https://webshare.leica-microsystems.com/latest/core/widefield/
- LAS X Core Confocal and FLIM viewers https://webshare.leica-microsystems.com/latest/core/confocal/
- Micro-Manager software for microscope control and image acquisition http://www.micro-manager.org
- GIMP http://www.gimp.org freeware for image adjustments, manipulations. Similar capabilities to Adobe Photoshop®.
- Scribus https://www.scribus.net/ page layout/publishing software for assembling multi-panel figures for publication, combines vector graphics and raster (images).
- Veusz https://veusz.github.io a scientific plotting and graphing program. For making publication-ready plots, provides full control of the plot parameters, appearance and size.
